Backyard Microbes
We are a group of microbiologists aiming to engage students from diverse communities into learning and discovering the fascinating microbial world by providing them the tools to find and study environmental microbes on their own.
Participant scientists and collaborator partners
Fernando Medina Ferrer (MBL Microbial Diversity alumni; UC Berkeley)
Diane Brache-Smith (MBL Microbial Diversity alumni; UC Merced)
Matthew Cope-Arguello (MBL Microbial Diversity alumni; UC Davis)
Virginia Russell (UC Berkeley)
Katie Shalvarjian (UC Berkeley)
Nina Abers (UC Berkeley)
Blake Downing (UC Berkeley)
Clare Gill (UC Berkeley)
Alienor Baskevitch (UC Berkeley)
Dinesh Gupta (UC Berkeley)
Jonathan Gropp (UC Berkeley)
Annie Choy (Hayward High School)
Matthew Nixon (Hayward High School)
Manvir Kaur Baidwan (Hayward High School)
Bharat Mehta (Tennyson High School)
Emerald Young (Early Academic Outreach Program, UC Berkeley)
Gina Saechao (Early Academic Outreach Program, UC Berkeley)
Overall Goals and Objectives
-
Introduce students from different High Schools with hands-on activities that will enable them to spot environmental microbes using (1) direct visualization (microscopy) and (2) an indirect detection technique that reveals the activity of the microbial enzyme urease.
- Provide each student with field microscopes and enzymatic activity tests that they could use with their families and friends at home and in regional trips, so they have the tools to independently discover the diversity of microbes living around us.
- Our long-term goal is to obtain student-generated data to map urease activity from diverse locations and microbial samples, and centralize this information on a website that will serve as a repository of field microbial activity. Students are thus able to participate in current research by discovering novel sites that will allow them to better understand our microbial world.
Background
Although our planet Earth is ruled by microbes, we are often not conscious about their role in our everyday environment. From past global climate changes to the oxygen that we now breathe, microbes are fundamental in our ecosystems, yet we usually only associate them with diseases. We aim to show students that the majority of microbes are not harmful, that we could find them virtually everywhere on Earth, and that their lifestyles have the capacity to modify the environment in which we live. Most importantly, we are yet not fully aware of the vast diversity of microbes and their in situ activities driving local changes. This opens an opportunity to study microbes and microbial activities in their native habitats, using simple and economical tools that could easily be deployed in the field by students themselves. Our goal is to give students the tools to investigate microbes, evocating the exploratory-driven research performed in the MBL Microbial Diversity course.
Activities
We developed two modular hands-on activities and encouraged students to perform their own discoveries by exploring environmental microbes and share their field findings.
Microscope visualization
An introduction of the concept of microscopy was followed by a hands-on activity where each student assembled a commercially available origami-based microscope (the Foldscope, ). Using this microscope, students observed environmental samples containing microbial biofilms, particularly filamentous green algae that are large and colorful candidates for visualization. Students were able to obtain pictures and share their findings, in addition to keep this portable microscope to observe samples of their own choice.

Enzymatic activity detection
Microbes continuously impact our world, even if we cannot see them. To visualize an example of microbially-induced change, students assembled a fast and simple colorimetric test to detect the activity of the enzyme urease in environmental microbial samples. This assay consists of two papers containing (a) the enzymatic substrate (urea), and (b) a pH indicator (phenol red), both assembled inside a microcentrifuge tube that serves as a closed reaction environment (). Upon introduction of a sample containing urease, a color change in the pH indicator from yellow to red due to the release of ammonia inside the tube allows to follow the enzymatic reaction according to the following equation:
Urea + Water -> Carbon Dioxide + Ammonia
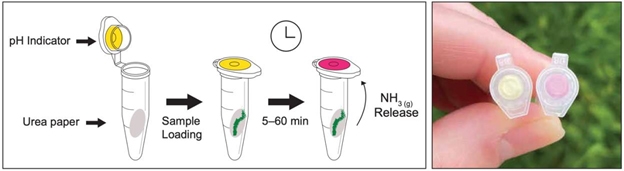
Assemblage of the assay is fast, simple, and it can be done without the need for a laboratory. The field test is easy to operate, and depending on the studentŌĆÖs grade, their data interpretation could range from a simple ŌĆśyes or noŌĆÖ answer (color change), to semi- quantification of the assay color change through time (representing enzyme kinetics). Students tested their assembled assays with different standards of urease that turn the indicator red within 5ŌĆō30 min, in addition to evaluate environmental biofilms for the presence of extracellular urease. All students were able to assemble as much enzymatic tests as wanted and keep them to test samples picked on their own after the activity. Learn more here.
╠²


Field testing of microbial samples
Equipped with the foldscope and the field activity assay, students were encouraged to independently explore their environment. From soil to microbial biofilms, outside their schools, homes and backyards, students could independently search for microbes and discover field urease activity. Students could also submit their field test results to the , where we aim to build the first environmental microbial urease activity database supported by field data from students that participated in this project. We hope that participation in this citizen science experience will make students aware of how scientific discoveries are conducted and further motivate them to engage in science.
Outcomes and Impacts
We worked together with Title I Schools from the East Bay in Northern California, so far reaching over 250 students from Hayward, Tennyson, Hercules, Pinole Valley, Richmond, and De Anza High Schools. We stablished a new collaboration network aimed to foster the knowledge in science and to motivate students in the pursue of higher education.
With this project we also started the development of an ever-growing geographical survey of environmental urease activity that is fed with student-generated data. This urease activity network map could be relevant for agriculture-impacted areas, serving as an indirect indicator of contaminated soils (being urea the major fertilizer used today). Thus, students could potentially engage in discoveries that directly affect their communities, such as the identification of nearby contaminated sites, or linking eutrophication of water bodies with urea contamination. The generation of data from field urease activity through this citizen science project also provides a useful resource to help find new and efficient urease-producing microbes, which could result of important applications in microbiology, biotechnology, and engineering.